Journal of Neurology, Neurological Science and Disorders
PCR-RFLP evidences peculiarities in Spinal Muscular Atrophy among Cuban Patients
Pita Rodríguez M1*, Zaldívar Vaillant T1, Zayas Guillot M1, Alvarez González MA1,2
2University of Havana
Cite this as
Rodríguez MP, Vaillant TZ, Guillot MZ, Alvarez González MA (2017) PCR-RFLP evidences peculiarities in Spinal Muscular Atrophy among Cuban Patients. J Neurol Neurol Sci Disord 3(1): 051-052. DOI: 10.17352/jnnsd.000021Introduction
Spinal Muscular Atrophy (SMA) is a lethal, autosomal recessive, neurodegenerative disorder characterized by progressive muscle weakness. SMA has an incidence of 1 in 6000-10000 live-births and a carrier frequency of 1:38–50 [1]. Previous reports describe genotype and frequency differences among ethnic groups [2,3]. In around 95% SMA results from the loss of SMN1 gene [4]. SMA can be classified into five clinical grades based on age of onset and severity. Cuba has a high degree of admixture [5], and previous studies in this population report a different SMN1homozygous deletion frequency [6], and skin color distribution of SMA I [2]. In this study, a molecular characterization of one hundred sixty-three patients was performed by PCR-RFLP methods regarding gender and skin color distribution.
Results
A molecular diagnosis of patients with clinical symptoms of SMA was performed by PCR-RFLP techniques described previously [7]. SMA classification did not consider type 0 and the type IV; four cases were excluded from this analysis. Skin color was self-reported by patients or parents during examination (W: White, M: Mestizo, B: Black).
The analysis of the 163 patients evidence a predominance of SMA type I (I:81, II:58, III:24).From this total, only 108 were positive for SMN1 exon 7 homozygous deletion (66.26 %).
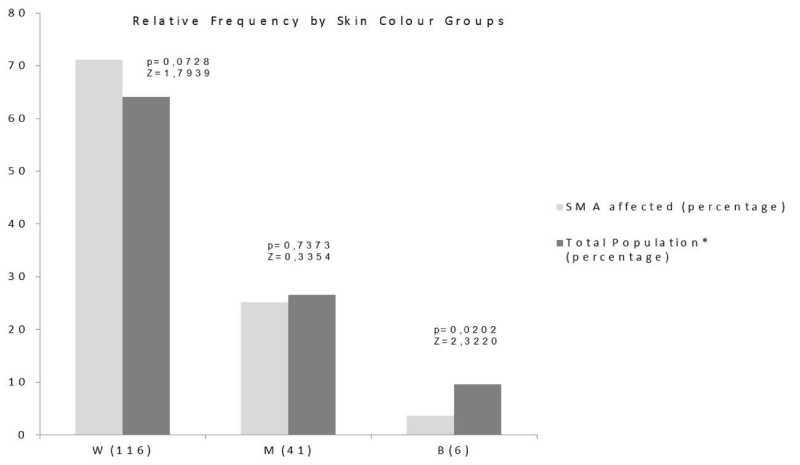
The analysis of relative frequency differences by skin color groups resulted significant for black patients exclusively (Figure 1).
Sex distribution showed a higher frequency in males (96 patients) compared to females (67 patients) (p=0.0013, ratio=1.43/1). The same was obtained when only molecularly confirmed cases were considered (67 and 41 cases respectively) (p=0.0005, ratio= 1.63/1).
No significant differences (χ2=0.81 p=0.66) were found among SMA types regarding presence or absence of SMN1 exon 7. Mean age (in months) for females (37.82) and males (47.10) did not show significant differences (t= -0.682551, p=0.50). Skin color did not either have significant effect in mean age (W=42.98, M=44.27, B=43.07) (F=0.0034, p=0.997).
Discussion
The low proportion of SMN1 exon 7 homozygous deletion in Cuban patients could be due to intragenic mutations, not involving gene deletion, widely spread in our population. Cuba has a high degree of admixture and different ethnical frequencies of alleles with multiple copies of SMN1 have been described [3]. These findingscould justify the frequency differences of SMA in specific ethnics groups. Probably, in our SMA population, alleles harbor multiple copies even with the alleged intragenic mutations.
Previous studies report SMA phenotype modifiers. Plastin 3 (PLS3) is a calcium dependent protein that co-localizes with SMN in granules throughout motor neuron axons. Overexpression of PLS 3 has been described in asymptomatic females with SMN1 homozygous deletion and it has been suggested that PLS3 may be a sex-specific SMA phenotype modifier [8]. This could be at least a partial explanation for the statistically significant differences in sex distribution in our patients.
Conclusion
We found a different SMN1 exon 7 homozygous deletion percentages for SMA Cuban patients and a significantly higher incidence in males compared to females. Additionally, we demonstrated a very low relative frequency in black skin color patients.
In summary, these findings among Cuban SMA Patients do demand for additional research on particular genetic bases of SMA in our population. These results have serious implications for molecular diagnostic and genetic counseling that can be offered to SMA patients and relatives.
Statistical analysis
Chi square test, Student´s t-test, proportions comparison test and one-way ANOVA were employed to analyze obtained data.
Ethics statement
Ethical guidelines of the Institute of Neurology and Neurosurgery were strictly followed.
Research limitations
A PCR-RFLP method only detects presence or absence of SMN1 gene. Heterozygosis condition for SMN1, intragenic mutations, hybrid genes and other sequence alterations cannot be detected. Further experiments like quantification of SMN1/SMN2 copy number and gene sequencing must be done to clarify these findings.
- Ben-Shachar S, Orr-Urtreger A, Bardugo E, Shomrat R, Yaron Y (2011) Large-scale population screening for spinal muscular atrophy: clinical implications. Genet. Med 13:110-114. Link: https://goo.gl/jtKfgS
- Zaldívar T, Montejo Y, Acevedo AM, Guerra R, Vargas J et al. (2005) Evidence of reduced frequency of spinal muscular atrophytypeIin the Cuban population. Neurology; 65: 636-638. Link: https://goo.gl/7xWQL4
- Hendrickson BC, Donohoe C, Akmaev VR, Sugarman EA, Labrousse P et al. (2009) Differences in SMN1allelefrequencies among ethnic groups within North America. J. Med.Genet 46: 641-644. Link: https://goo.gl/tvTfof
- Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P et al. (1995) Identification and characterization of a spinal muscularatrophy-determininggene. Cell 80:155-165. Link: https://goo.gl/1aGTek
- Marcheco-Teruel B, Parra EJ, Fuentes-Smith E, Salas A, Buttenschøn HN, et al. (2014) Cuba: Exploring the History of Admixture and the Genetic Basis of Pigmentation Using Autosomal and Uniparental Markers. PLoS Genet 10: e1004488. Link: https://goo.gl/BcWtwT
- Viñas CI, Martín I, Zaldívar T, Garofalo N, Zayas M et al. (2013) Análisisgenético molecular en Atrofia Muscular Espinal. Rev. Chil. Pediatr.; 84: 499-504. Link: https://goo.gl/NKkpzv
- Van der Steege G, Grootscholten PM, van der Vlies P, Draaijers TG, Osinga J et al. (1995) PCR-based DNA test to confirm clinical diagnosis of autosomal recessive spinal muscular atrophy. Lancet 345: 985-986. Link: https://goo.gl/D13Sdg
- Oprea GE, Kröber S, McWhorter ML, Rossoll W, Müller S et al. (2008) Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science 320: 524-527. Link: https://goo.gl/k94whq
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
